2024
-
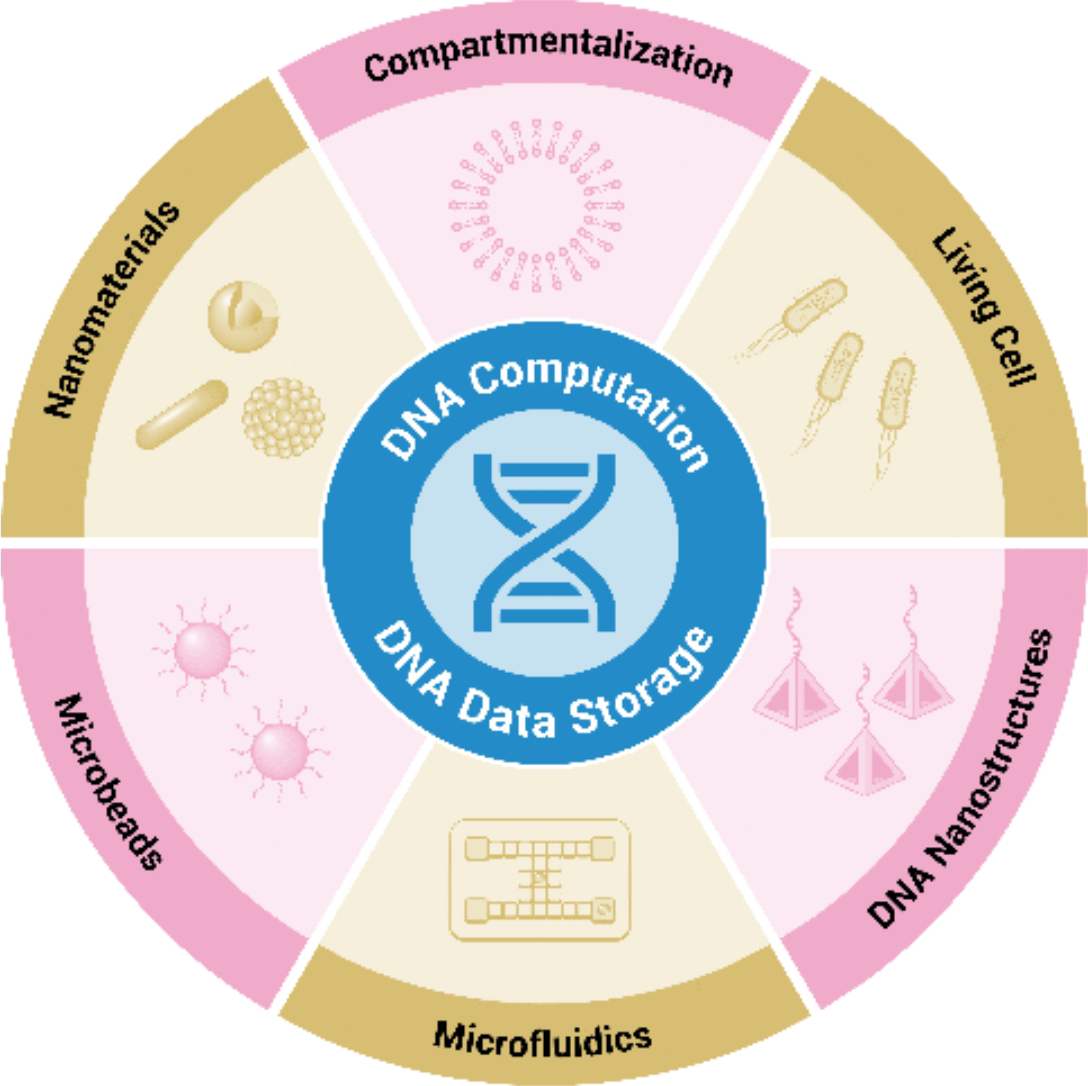
315..Kunjie Li†, Heng Chen†, Dayang Li†, Chaoyong Yang, Huimin Zhang* and Zhi Zhu*. Empowering DNA-Based Information Processing: Computation and Data Storage. ACS Appl Mater Interfaces. 2024, 16, 50, 68749–68771.
-
314.Xing Xu†, Qianxi Wen†, Tianchen Lan, Liuqing Zeng, Yonghao Zeng, Shiyan Lin, Minghao Qiu, Xing Na and Chaoyong Yang*. Time-resolved single-cell transcriptomic sequencing. Chem Sci. 2024,15,19225-19246.

-
313.Lianyu Lu, Yaohui Wang, Yue Ding, Yuqing Wang, Zhi Zhu, Jinsong Lu*, Liu Yang*, Peng Zhang*, Chaoyong Yang*. Profiling Phenotypic Heterogeneity of Circulating Tumor Cells through Spatially Resolved Immunocapture on Nanoporous Micropillar Arrays. ACS Nano. 2024, 18, 31135–31147.

-
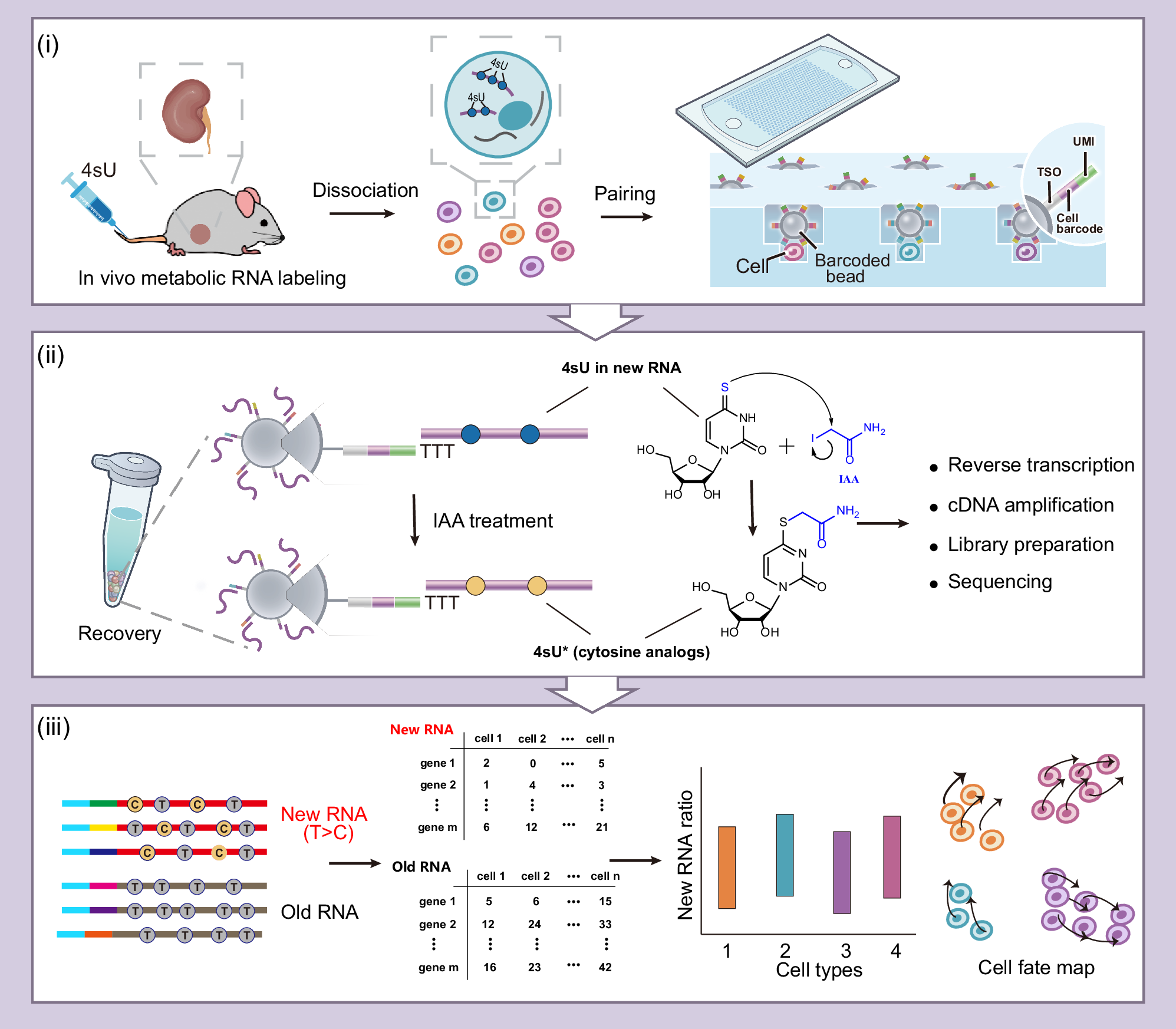
312.Kun Yin†, Yiling Xu†, Ye Guo†, Zhong Zheng†, Xinrui Lin, Meijuan Zhao, He Dong, Dianyi Liang, Zhi Zhu, Junhua Zheng*, Shichao Lin*, Jia Song*, Chaoyong Yang*. Dyna-vivo-seq Unveils Cellular RNA Dynamics during Acute Kidney Injury via in vivo Metabolic RNA Labeling-based scRNA-seq. Nat. Commun. 2024, 15, 9866.
-
311.Shanshan Liang,Chong Li,Yu Ning,Rui Su,Mingyin Li,Yihao Huang,Yuning Zou,Liu Yang*, Xing Xu*,and Chaoyong Yang*. DMF-Bimol: Counting mRNA and Protein Molecules in Single Cells with Digital Microfluidics. Analytical chemistry, 2024, 96, 17253–17261.

-
310.Mingyin Li⊥, Xing Na⊥, Fanghe Lin, Shanshan Liang, Yuehan Huang, Jia Song*, Xing Xu*,and Chaoyong Yang*. DMF-ChIP-seq for Highly Sensitive and Integrated Epigenomic Profiling of Low-Input Cells. ACS Appl Mater Interfaces. 2024, 16, 52047-52058.
-
309.Xi Zeng#, Xiaoping Yang#, Zhixing Zhong#, Xin Lin, Qiuyue Chen, Shaowei Jiang, Mengwu Mo,Shichao Lin, Huimin Zhang, Zhi Zhu*, Jin Li*, Jia Song*, and Chaoyong Yang*. AMAR-seq: Automated Multimodal Sequencing of DNA Methylation, Chromatin Accessibility, and RNA Expression with Single-Cell Resolution. Anal Chem. 2024, 96, 15295-15303.
-
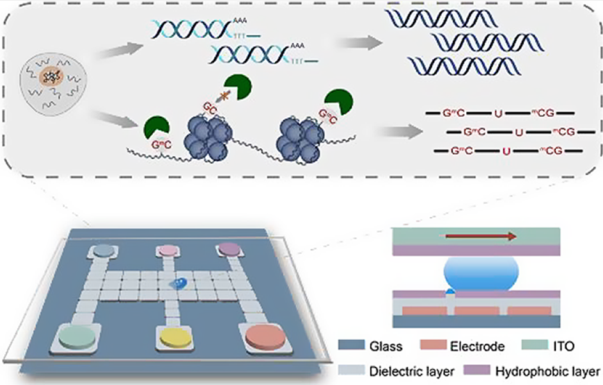
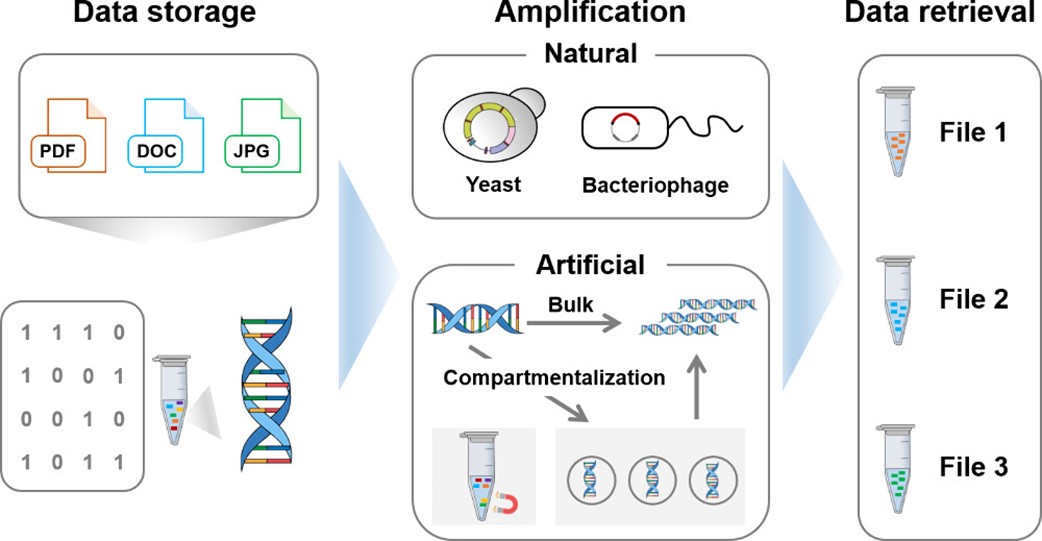
308.JialuZhang, MingyingChen, GuihongLin, SinongLiu, ChaoyongYang, and YanlingSong*.Advanced DNA Amplification for Efficient Data Storage.ACS Appl Mater Inter. 2024, 16, 48870–48879.
-
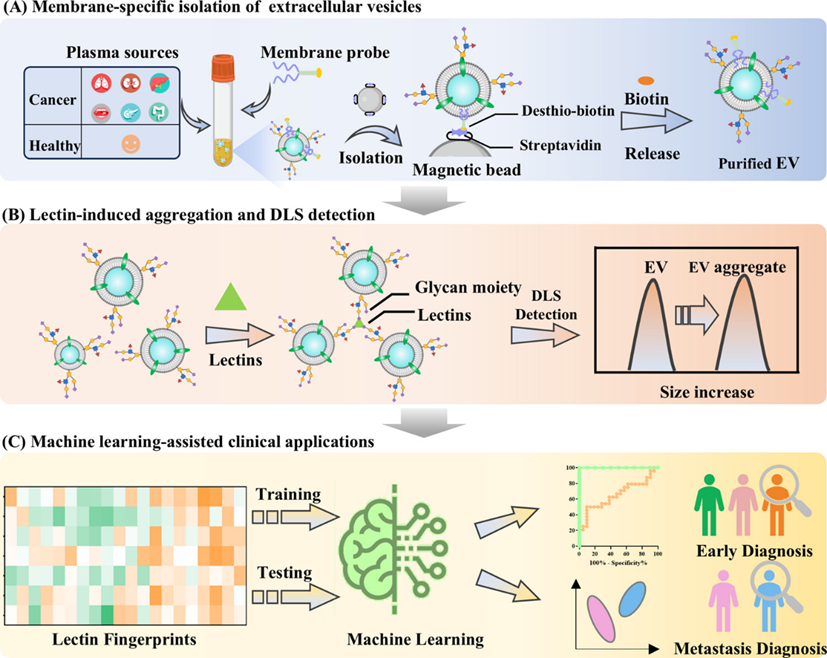
307.Guihua Zhang, Xiaodan Huang, Yanli Gong, Yue Ding, Hua Wang, Huimin Zhang, Lingling Wu, Rui Su, Chaoyong Yang and Zhi Zhu*. Fingerprint Profiling of Glycans on Extracellular Vesicles via Lectin-Induced Aggregation Strategy for Precise Cancer Diagnostics. J Am Chem Soc. 2024,146, 42, 29053–29063.
-
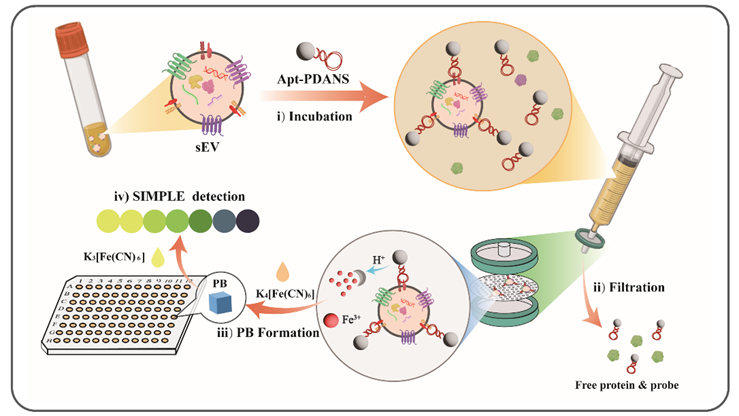
306.GuihuaZhang,QiannanZhang,HuanghuangZhu,RuiMa,XiaodanHuang,ShiyunCen,ChaoyongYang,RuiSu*,andZhiZhu*.Fast Isolation and Sensitive Multicolor Visual Detection of Small Exracellular Vesicles by Multifunctional Polydopamine Nanospheres.Anal Chem. 2024, 96, 15177-15184.
-
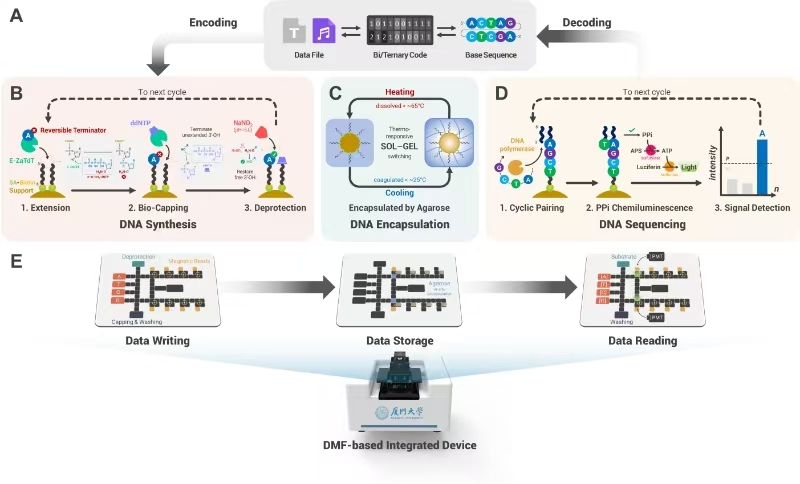
305.Kunjie Li, Xiaoyun Lu, Jiaqi Liao, Heng Chen, Wei Lin, Yuhan Zhao, Dongbao Tang, Congyu Li, Zhenyang Tian, Zhi Zhu, Huifeng Jiang, Jun Sun*, Huimin Zhang* and Chaoyong Yang*. DNA-DISK: automated end-to-end data storage via enzymatic single-nucleotide DNA synthesis and sequencing on digital microfluidics. PNAS. 2024, 121, e2410164121.
-
304.Junnan Guo, Di Sun, Kunjie Li, Qi Dai, Shichen Geng, Yuanyuan Yang, Mengwu Mo, Zhi Zhu, Chen Shao, Wei Wang, Jia Song, Chaoyong Yang, and Huimin Zhang*.Metabolic Labeling and Digital Microfluidic Single-Cell Sequencing for Single Bacterial Genotypic-Phenotypic Analysis. Small. 2024, 11, 2402177.

-
303.Kun Yin+, Meijuan Zhao+, Yiling Xu+, Zhong Zheng+, Shanqing Huang, Dianyi Liang, He Dong, Ye Guo,Li Lin, Jia Song, Huimin Zhang, Junhua Zheng*, Zhi Zhu*, and Chaoyong Yang*. Well-Paired-Seq2: High-Throughput and High-Sensitivity Strategy for Characterizing Low RNA-Content Cell/Nucleus Transcriptomes. Anal Chem. 2024, 96, 6301-6310.

-
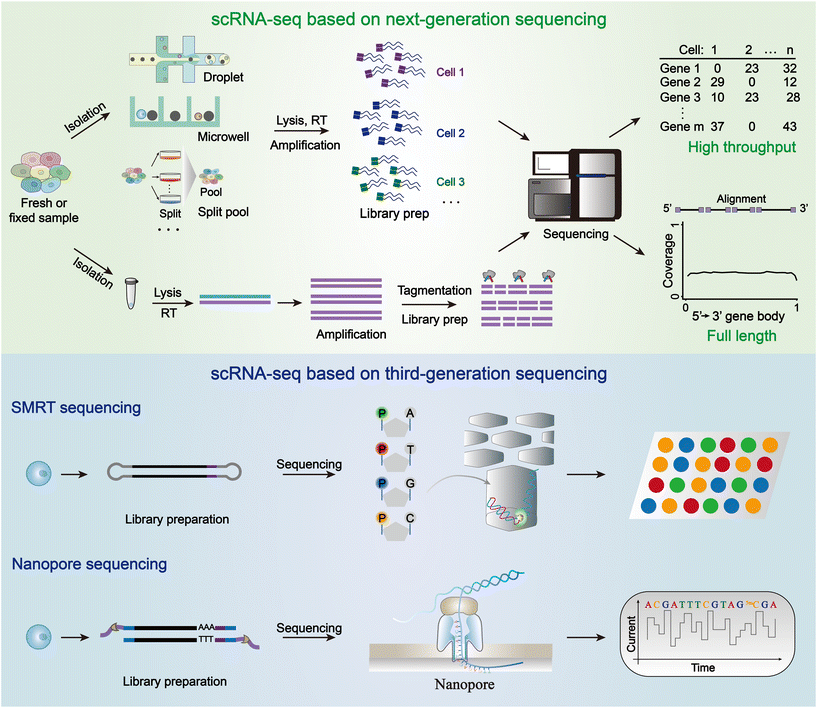
302.Shanqing Huang, Weixiong Shi, Shiyu Li, Qian Fan, Chaoyong Yang, Jiao Cao* and Lingling Wu*. Advanced sequencing-based high-throughput and long-read single-cell transcriptome analysis. Lab Chip. 2024, 24, 2601-2621.
-
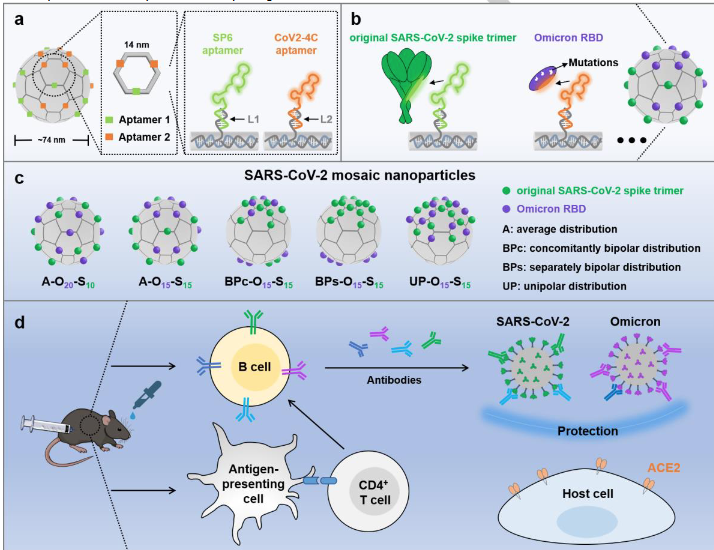
301.Jialu Zhang+, Yunyun Xu+, Mingying Chen, Shengwen Wang,Guihong Lin,Yihao Huang,Chaoyong Yang, Yang Yang*, and Yanling Song*. Spatial Engineering of Heterotypic Antigens on a DNA Framework for the Preparation of Mosaic Nanoparticle Vaccines with Enhanced Immune Activation against SARS-CoV-2 Variants. Angew Chem Int Ed Engl. 2024, e202412294.
-
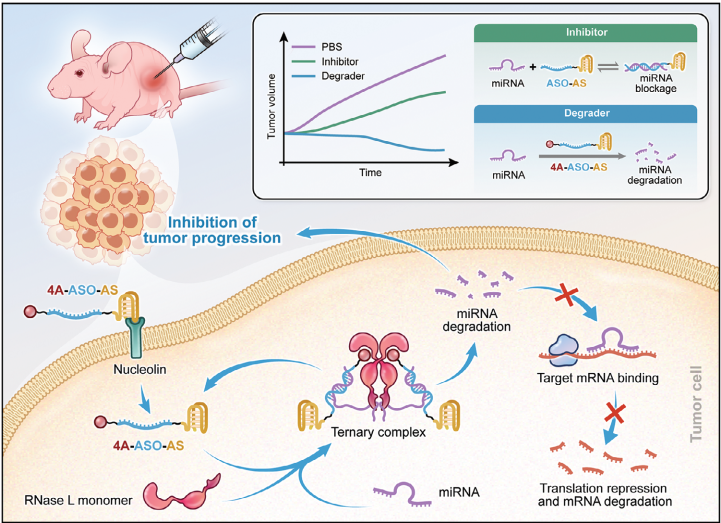
300.Yuan Fang, Qiuyue Wu, Feiyu Wang, Ye Liu, Huimin Zhang, Chaoyong Yang,* and Zhi Zhu*. Aptamer-RIBOTAC Strategy Enabling Tumor-Specific Targeted Degradation of MicroRNA for Precise Cancer Therapy. Small Methods, 2024, 2400349.
-
299.Yingwen Chen, Xuanqun Wang, Xing Na, Yingkun Zhang, Linfeng Cai, Jia Song, and Chaoyong Yang*. DMF-DM-seq: Digital-Microfluidics Enabled Dual-Modality Sequencing of Single-Cell mRNA and microRNA with High Integration, Sensitivity, and Automation. Anal Chem. 2024, 96, 12916-12926.

-
298.Weizhi Liu+, Shanqing Huang+, Ye Guo, Xingrui Li, He Dong, Juan Li, Chaoyong Yang, Zhi Zhu. Deciphering molecular response of cell-cell interactions at the single-cell level by precise on-demand cell assembly. Sci Bull (Beijing). 2024, 69, 2342-2345.

-
297.Weixiong Shi1, Jing Zhang1, Shanqing Huang, Qian Fan, Jiao Cao, Jun Zeng, Lingling Wu,* Chaoyong Yang*. Next-Generation Sequencing-Based Spatial Transcriptomics: A Perspective from Barcoding Chemistry. JACS Au. 2024, 4, 1723–1743
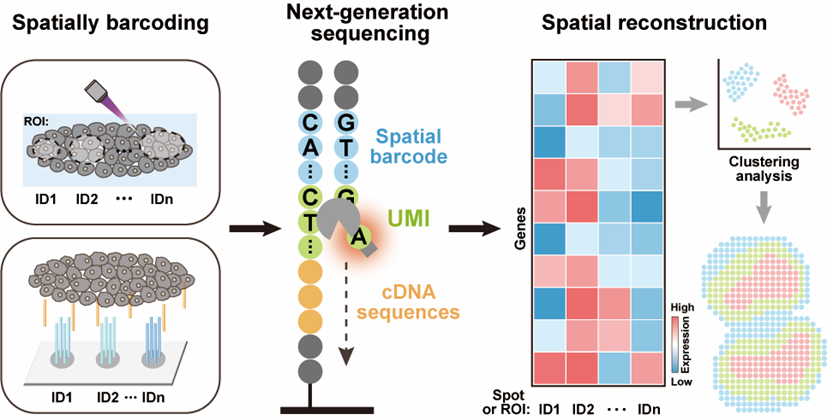
-
296.Linfeng Cai, Li Lin, Shiyan Lin, Xuanqun Wang, Yingwen Chen, Huanghuang Zhu, Zhi Zhu, Liu Yang, Xing Xu, Chaoyong Yang. Highly Multiplexing, Throughput and Efficient Single‐Cell Protein Analysis with Digital Microfluidics. Small Methods, 2024, doi: https://doi.org/10.1002/smtd.202400375
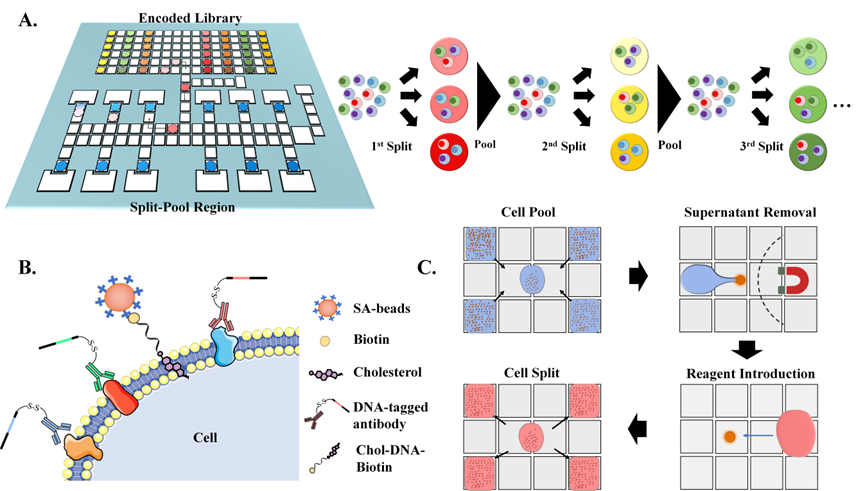
-
295.Xing Xu, Xi Zeng, Xin Lin, Shiyan Lin, Shanshan Liang, Tian Tian, Rui Su, Jia Song, Chaoyong Yang. DMF-scMT-seq Linking Methylome and Transcriptome within Single Cells with Digital Microfluidics. Science China Chemistry. 2024, 67, 2070–2078.
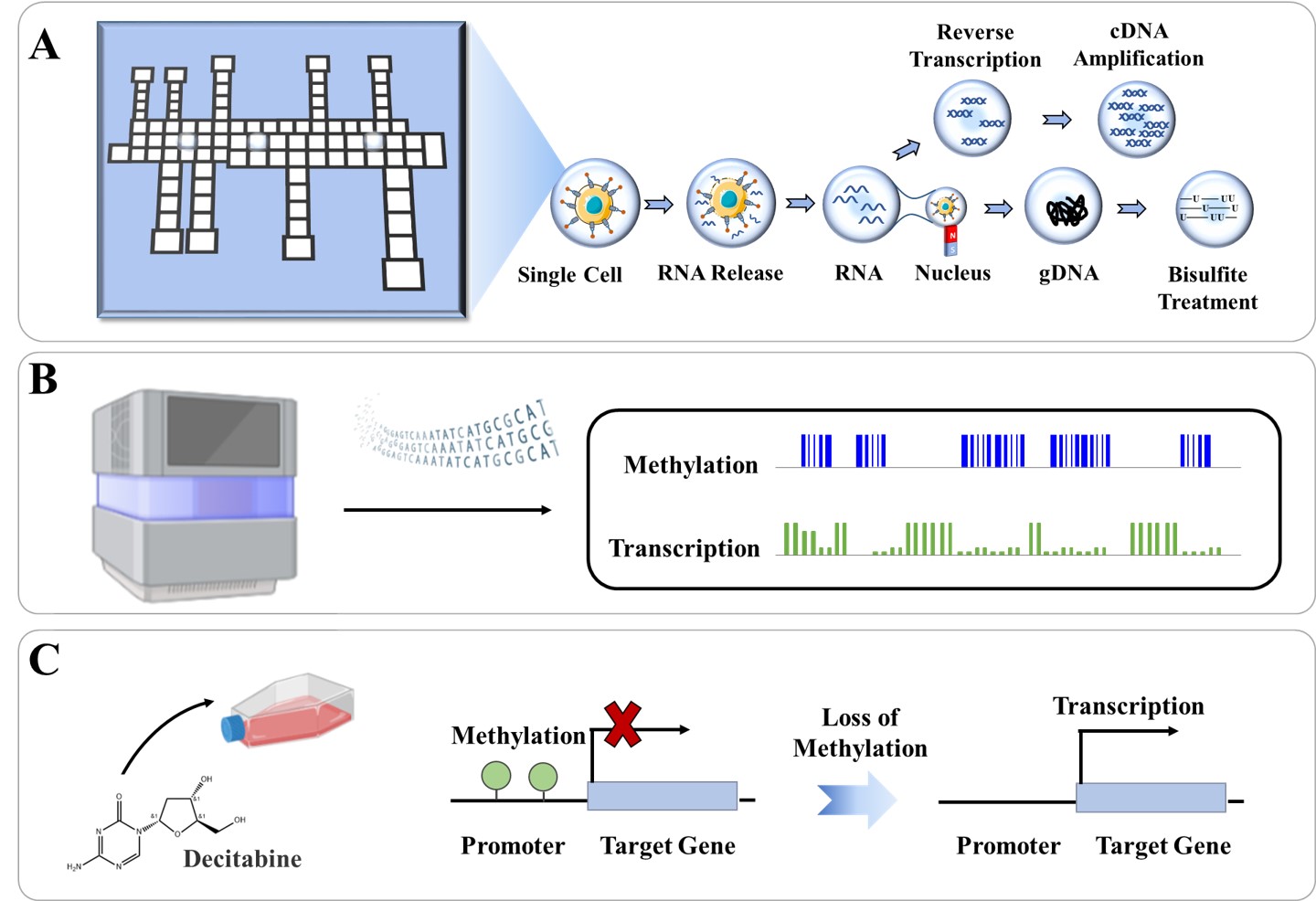
-
294.Yu An, Jinchao Zhu, Qihui Xie, Jianzhou Feng, Yanli Gong, Qian Fan, Jiao Cao, Zhi Huang, Weixiong Shi, Qingyuan Lin, Lingling Wu, Chaoyong Yang, Tianhai Ji. Tumor Exosomal ENPP1 Hydrolyzes cGAMP to Inhibit cGAS-STING Signaling. Adv Sci. 2024, 11, 2308131.
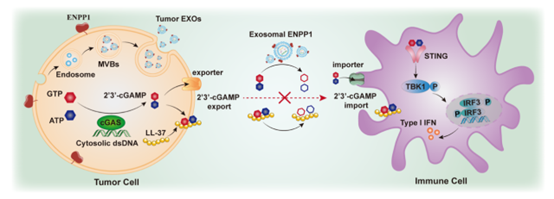
-
293.Jiao Cao, Zhong Zheng, Di Sun, Xin Chen, Rui Cheng, Tianpeng Lv, Yu An, Junhua Zheng, Jia Song, Lingling Wu & Chaoyong Yang. Decoder-seq enhances mRNA capture efficiency in spatial RNA sequencing. Nat Biotechnol. 2024,42,1735–1746.
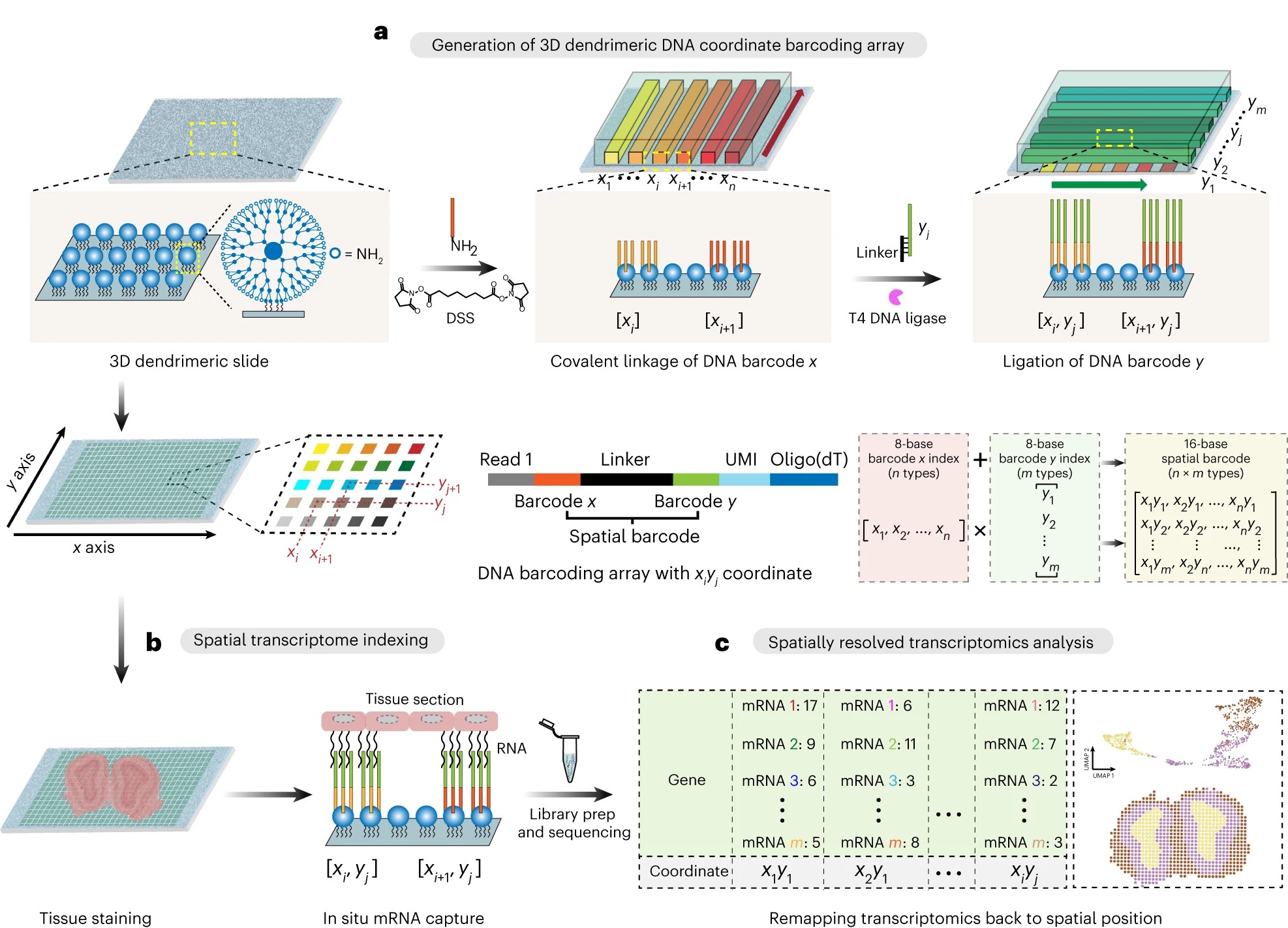
-
292.Zhong Z, Hou J, Yao Z, Dong L, Liu F, Yue J, Wu T, Zheng J, Ouyang G, Yang C, Song J. Domain generalization enables general cancer cell annotation in single-cell and spatial transcriptomics. Nat Commun. 2024, 15, 1929.
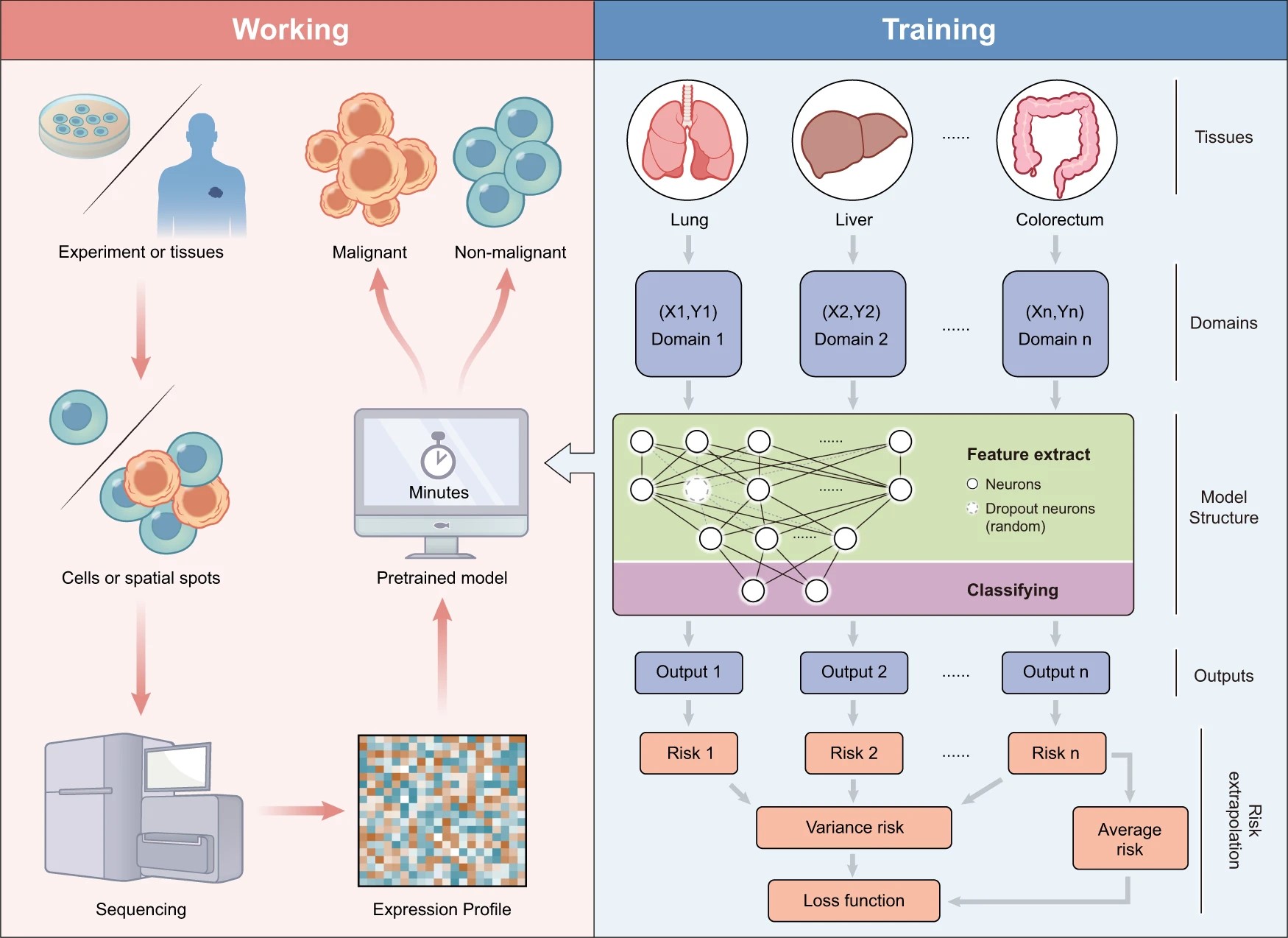
-
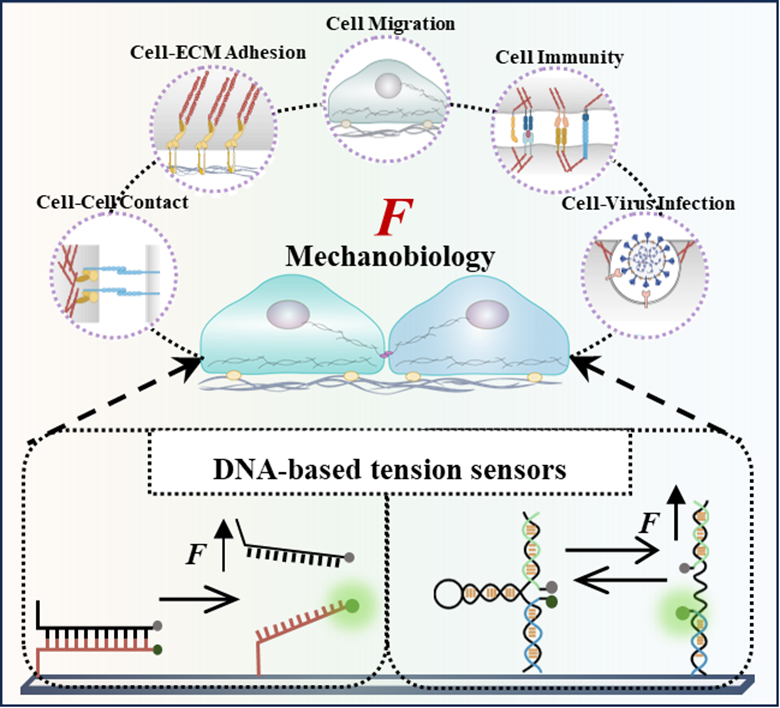
291.Yihao Huang, Ting Chen, Xiaodie Chen, Ximing Chen, Jialu Zhang, Sinong Liu, Menghao Lu, Chong Chen, Xiangyu Ding, Chaoyong Yang, Ruiyun Huang, Yanling Song. Decoding Biomechanical Cues Based on DNA Sensors. Small. 2024, 20, 2310330.