Department of Chemical Biology
Yang's Lab,Department of Chemical Biology, Xiamen University
Professor Yang
 Know more>>
Professor
Department of Chemical Biology, Xiamen University
Know more>>
Professor
Department of Chemical Biology, Xiamen University
Contact Information
Yang's LaboratoryRoom 532, Lujiaxi Building, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China
Ph: +86 (0) 592-218 7601cyyang@xmu.edu.cn
Manuscript by Xing Xu et al. has been accepted by Anal. Chem.
2020-05-19 14:47:08

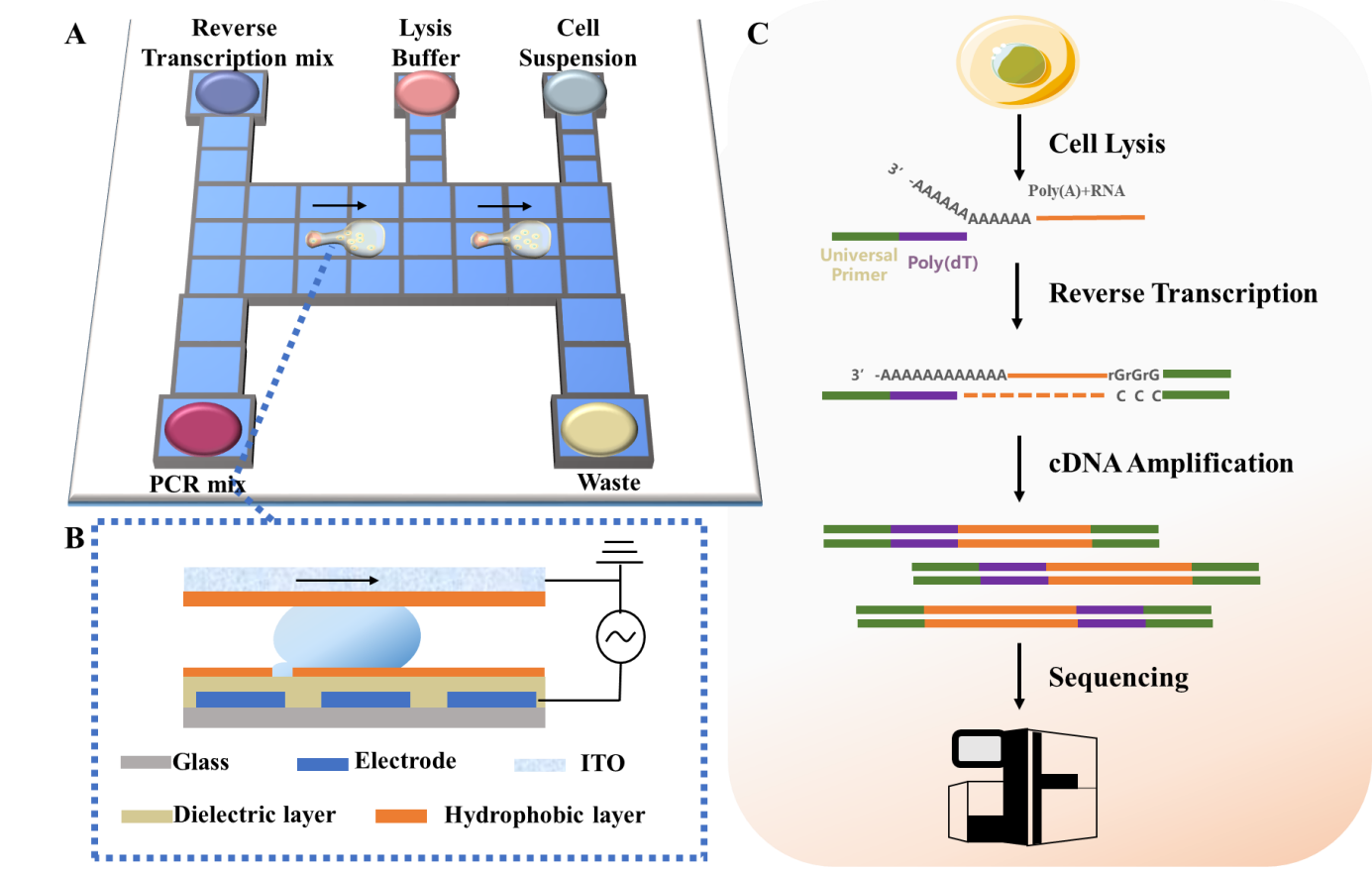
Single-cell RNA sequencing (scRNA-seq) is a powerful method in investigating single-cell heterogeneity to reveal rare cells, identify cell subpopulations and construct a cell atlas. Conventional bench-top methods for scRNA-seq including multi-step operations, are labor intensive, reaction inefficient, contamination prone and reagent consuming. Here we report a digital microfluidics-based single-cell RNA sequencing (Digital-RNA-seq) for simple, efficient and low-cost single-cell mRNA measurements. Digital-RNA-seq automates fluid handling as discrete droplets to sequentially perform protocols of scRNA-seq. To overcome the current problems of single-cell isolation in efficiency, integrity, selectivity and flexibility, we propose a new strategy, passive dispensing method, relying on well-designed hydrophilic-hydrophobic microfeatures to rapidly generate single-cell sub-droplets when a droplet of cell suspension is encountered. For sufficient cDNA generation and amplification, Digital-RNA-seq uses nanoliter reaction volumes and hydrophobic reaction interfaces, achieving high sensitivity in gene detection. Additionally, the stable droplet handling and oil-closed reaction space featured in Digital-RNA-seq ensure highly accurate measurement. We demonstrate the functionality of Digital-RNA-seq by quantifying heterogeneity among single cells, where Digital-RNA-seq shows excellent performance in rare transcript detection, cell type differentiation and essential gene identification. With the advantages of automation, sensitivity, and accuracy, Digital-RNA-seq represents a promising scRNA-seq platform for a wide variety of biological applications.