Professor Yang
 Know more>>
Professor
Department of Chemical Biology, Xiamen University
Know more>>
Professor
Department of Chemical Biology, Xiamen University
Contact Information
Yang's LaboratoryRoom 532, Lujiaxi Building, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China
Ph: +86 (0) 592-218 7601cyyang@xmu.edu.cn
Liyuan Lin's paper has been accepted by Acc. Chem. Res.
2021-04-02 22:37:00

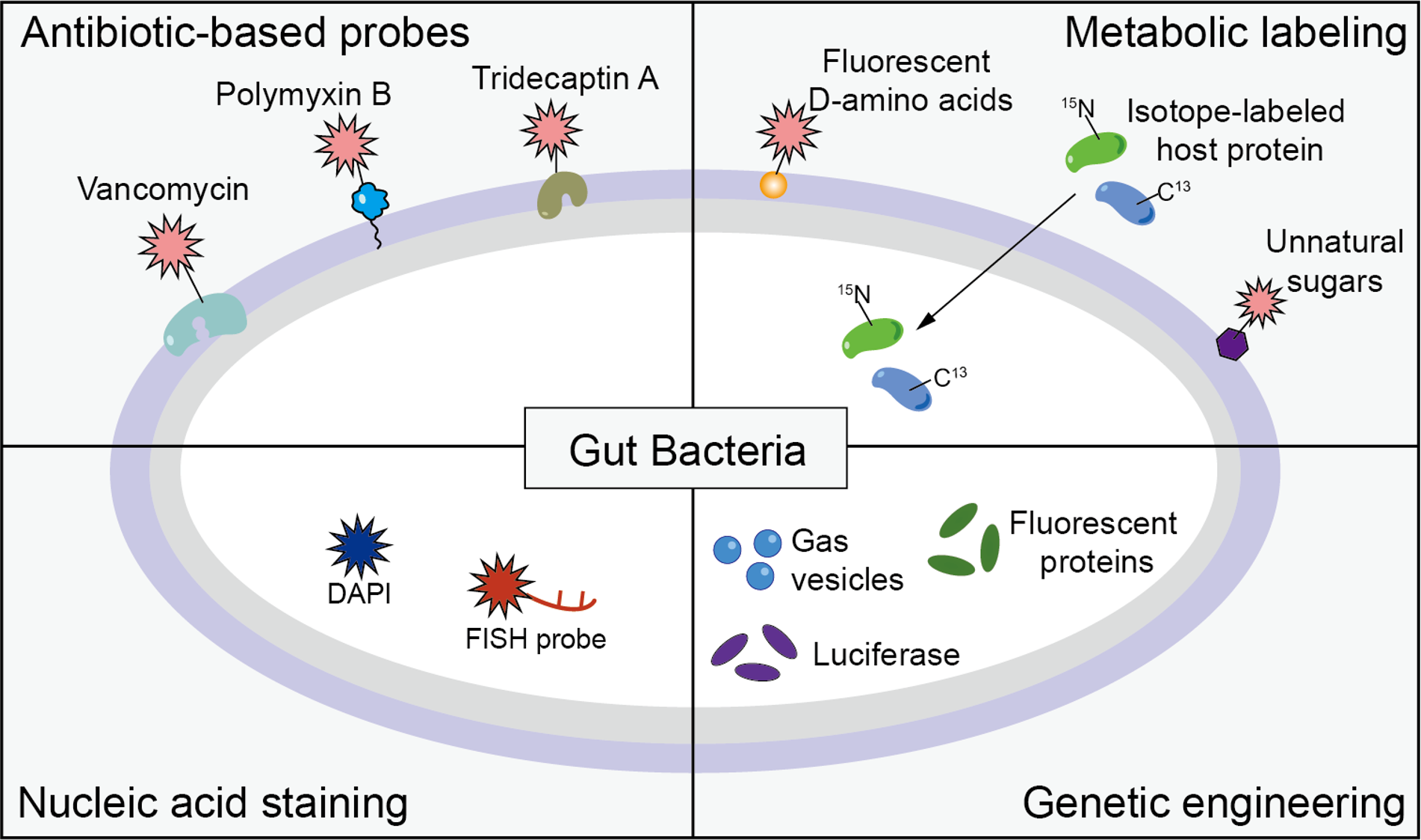
As a newly discovered organ, gut microbiotas have been extensively studied in the last two decades, with their highly diverse and fundamental roles in the physiology of many organs and systems of the host being gradually revealed. However, most of the current research heavily relies on DNA sequencing-based methodologies. To truly understand the complex physiological and pathological functions played by commensal and pathogenic gut bacteria, we need more powerful methods and tools, among which imaging strategies suitable for approaching this ecosystem in different settings are one of the most desirables. Although the phrase gut “dark matter” is often used in referring to the unculturability of many gut bacteria, it’s also applicable to describing the formidable difficulties in visualizing these microbes in the gut. To develop suitable and versatile chemical/biological tools for imaging bacteria in the gut, great efforts have been devoted in the past several years.
In this Account, we highlight the recent progresses made by our group and other labs in the development of visualization strategies for commensal microbiota and pathogenic bacteria in the gut. First, we summarize our efforts towards the development of antibiotic-derivatized staining probes that directly bind to specific bacterial surface structures for selective labeling of different groups of gut bacteria. Next, metabolic labeling-based imaging strategies, using unnatural amino acids, unnatural sugars, and stable isotopes, for imaging gut bacteria on various scales and in different settings are discussed in details. We then introduce the nucleic acid staining-based bacterial imaging, using either general nucleic acid-binding reagents or selective-labeling technique (e.g., fluorescence in situ hybridization) to meet the diverse needs in gut microbiota research. This classical imaging strategy has witnessed a renaissance in gut microbiota research owing to a series of new technical advancements. Furthermore, despite the notorious difficulties of performing genetic manipulations in many commensal gut bacteria, great endeavor has been made recently in engineering gut bacteria with reporters like fluorescent proteins and acoustic response proteins.
Our perspectives on the current limitations of the chemical tools/strategies and the future directions for improvement are also presented. We hope that this Account can offer valuable references to spark new ideas and invite new efforts to help decipher the complex biological and chemical interactions between commensal microbiota/pathogenic bacteria and the hosts.